Marker Choice
What is ALARMcoding?
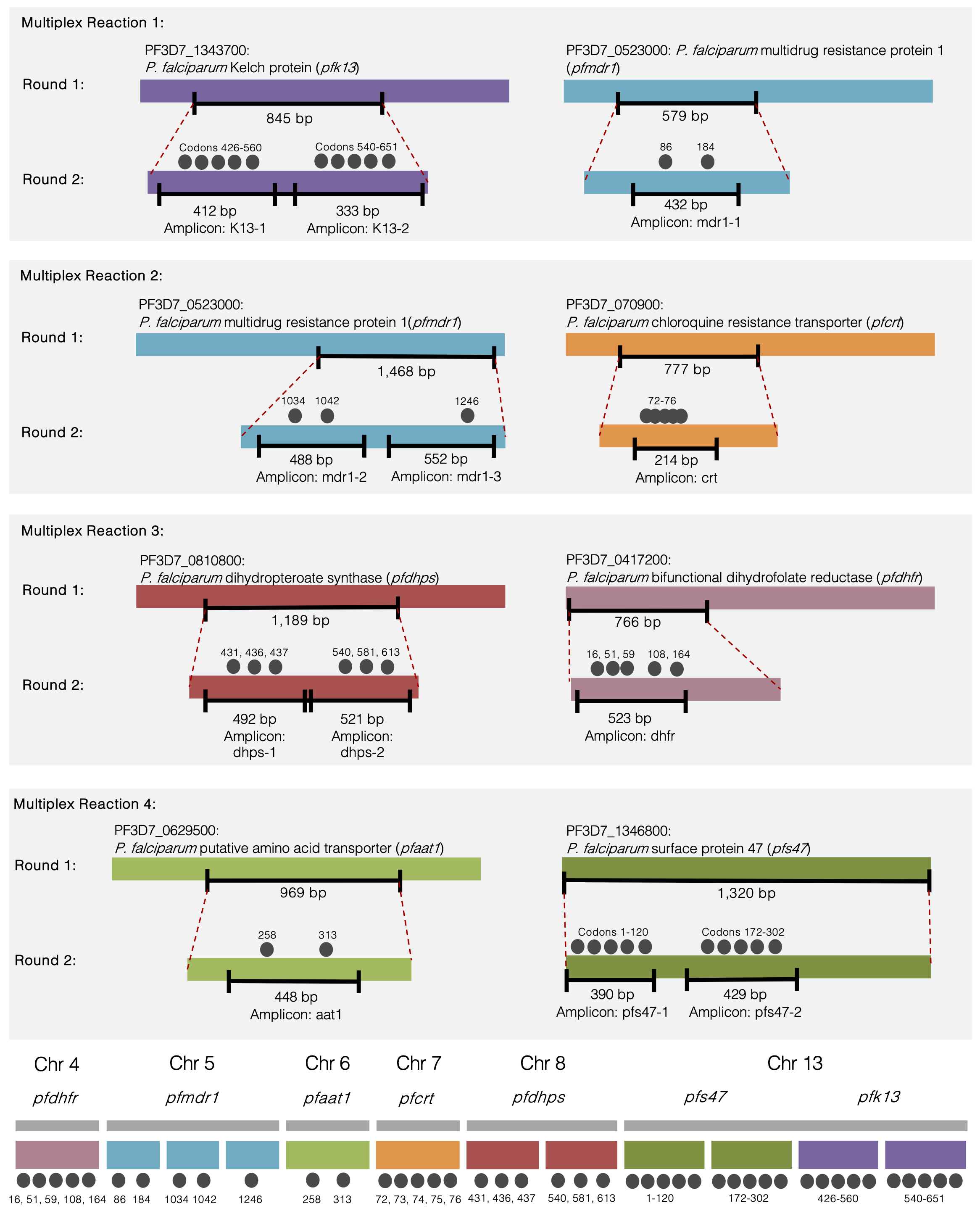
ALARMcoding, or Adaptive Loci of Antimalarial Resistance Markers coding, is a multiplex and nested PCR methodology to amplify 6 markers of drug resistance and one marker of vector competence. This panel has been optimised for asymptomatic data but can be used across a range of parasite densities, including clinical infections, and uses Amplicon Sequencing technology to also identify codons of interest in both single- and multiple-clone infections, where the latter are common in highly endemic countries.
Drug Resistance Markers
pfcrt, dhfr, dhps, K13, mdr1 and aat1 are molecular markers of drug resistance in Plasmodium falciparum. CQR is caused by common mutants in genes that encode P. falciparum transmembrane proteins chloroquine resistance transporter (PfCRT, codons 72-76), an ATP-binding cassette transporter and homologue of multidrug-resistance P-glycoprotein (PfMDR1 or Pgh1, codons 86, 184, 1034, 1042, 1246) and a putative amino acid transporter (PfAat1, codons 258, 313). For a detailed description on these markers see this page.
Vector Competence: A New Strategy
To address the recent invasion of An. stephensi in Africa, a marker of vector competence was included in the ALARMcoding panel. P. falciparum surface protein 47 (pfs47) encodes for the 6-cysteine protein pfs47 (also known as P47 or TEP1) that is expressed on the gametocyte surface and allows region-specific P. falciparum to evade the local vector immune response in the ookinete stage and persist to later developmental stages. This mediates the extent to which P. falciparum survives. pfs47-mediated immune evasion in the Anopheles spp. vector is required for parasites to persist in an endemic area (Molina-Cruz et al. 2013). There is strong geographic structure of pfs47 with lower haplotype diversity out of Africa (Anthony et al. 2007; Molina-Cruz, Canepa, et al. 2015; Molina-Cruz, Zilversmit, et al. 2015), supporting observations that:
- P. falciparum isolates from Africa could infect both An. gambiae (African vector) and An. stephensi (South/Southeast Asian vector), while P. falciparum isolates from southeast Asia had a lower success at infecting An. gambiae (Hume, Lyons, and Day 2003), and
- African strains (GB4 and NF54) were able to survive in An. gambiae but 7G8 (New World strain) cannot (Canepa, Molina-Cruz, and Barillas‐Mury 2016).
In the context of malaria genomic surveillance of drug resistance, pfs47 (chromosome 13) can be monitored to inform whether parasites have migrated/invaded from another region. It can therefore also be used as a positive control to confirm the vector-parasite relationship in the region of interest. Non-synonymous mutations in T236I, V247A, S242L and I248L resulted in the mosquito immune system clearing the parasite (Canepa, Molina-Cruz, and Barillas‐Mury 2016). Polymorphisms at codons 236 and 242 were found to be strongly related with geographic variation in the pfs47 gene across South America, Africa, Asia, and the Pacific (Molina-Cruz et al. 2021).
A recent study characterised polymorphisms of entire pfs47 gene in Western Kenya and found low and non-significant FST, with 8 loci that segregated in the population: E27D, E188D, P194H, L240I, I249L, N272I/Y, I304L. Only P194H and N272I were identified as SNPs while the other loci were parsimony informative (Onyango et al. 2021).