| Marker name | Length (base pairs, bp) | Expected position on gel (bp) | Diagnostic codons |
|---|---|---|---|
| K13-in-1 | 412 bp + MID (10 bp) | 422 | 426-560 |
| K13-in-2 | 333 bp + MID (10 bp) | 343 | 540-651 |
| mdr1-in-1 | 432 bp + MID (10 bp) | 442 | 86, 184 |
| mdr1-in-2 | 488 bp + MID (10 bp) | 498 | 1034, 1042 |
| mdr1-in-3 | 522 bp + MID (10 bp) | 532 | 1246 |
| crt-in | 214 bp + MID (10 bp) | 224 | 72-76 |
| dhfr-in | 523 bp + MID (10 bp) | 533 | 16, 51, 59, 108, 164 |
| dhps-in-1 | 492 bp + MID (10 bp) | 502 | 431, 436, 437 |
| dhps-in-2 | 521 bp + MID (10 bp) | 531 | 540, 581, 613 |
| aat1-in | 448 bp + MID (10 bp) | 458 | 258, 313 |
| pfs47-in-1 | 390 bp + MID (10 bp) | 400 | 1-120 |
| pfs47-in-2 | 429 bp + MID (10 bp) | 439 | 172-302 |
PCR: Important Notes
PCR General Workflow
This two-step protocol requires first, the amplification of the “outer” fragment and second, the amplification of the “inner” fragment. You can set up your plates with either 32 or 48 samples at a time. Handling more samples is not advisable, as it may lead to longer processing times and an increased risk of human error when adding gDNA. An example of conducting the PCR with 32 samples at a time is provided below.
For Step 1:
Each master mix is exactly the same.
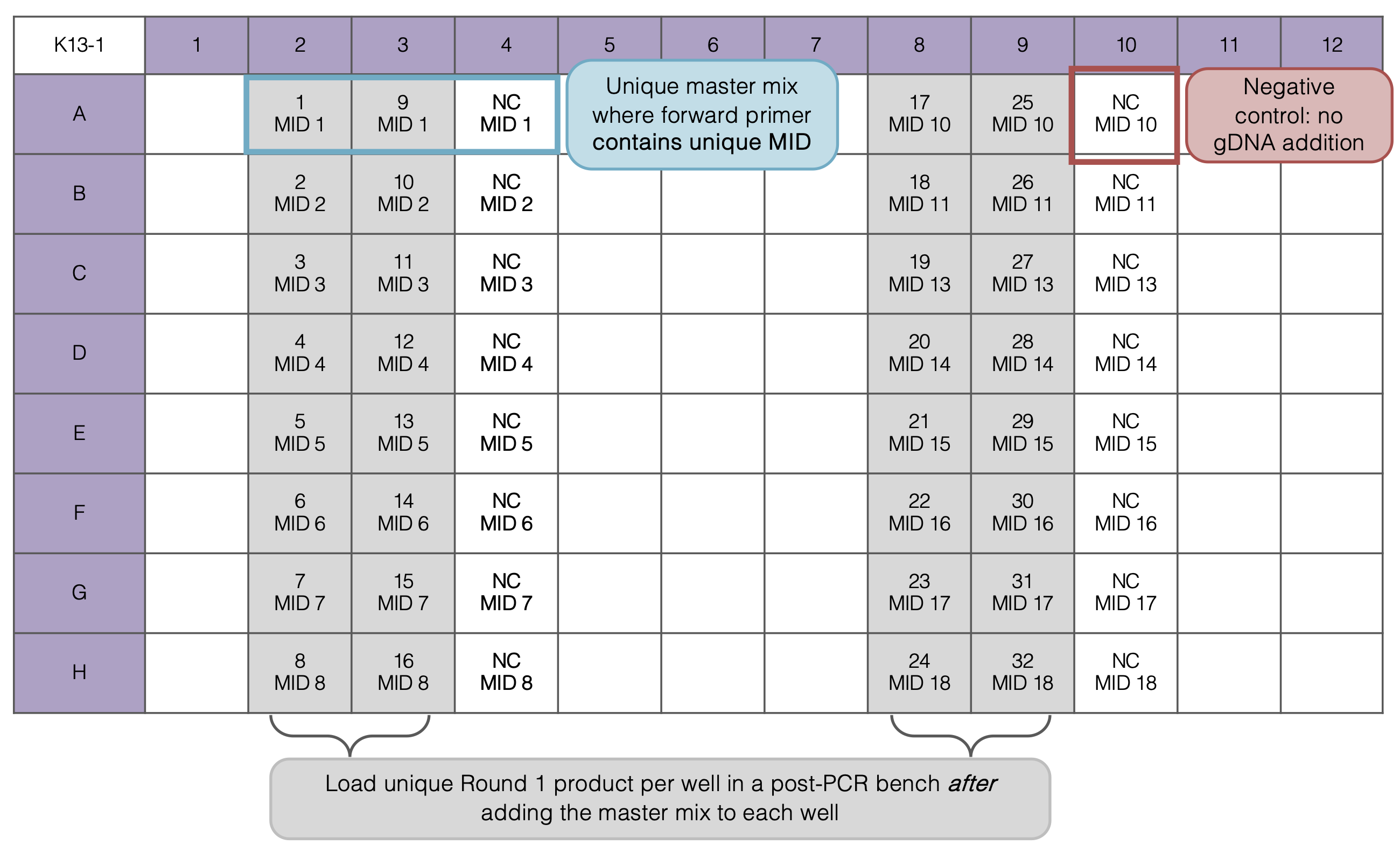
For Step 2:
Master mix differ by each row - corresponding to the molecular identifier (MID) tag on the forward primer, the reverse primer remains the same for each master mix. You will need to order unique MIDs + forward primer per molecular marker for each set of isolates you plan to pool together. In this work, I used 110 MIDs. See the list of MID tags here.
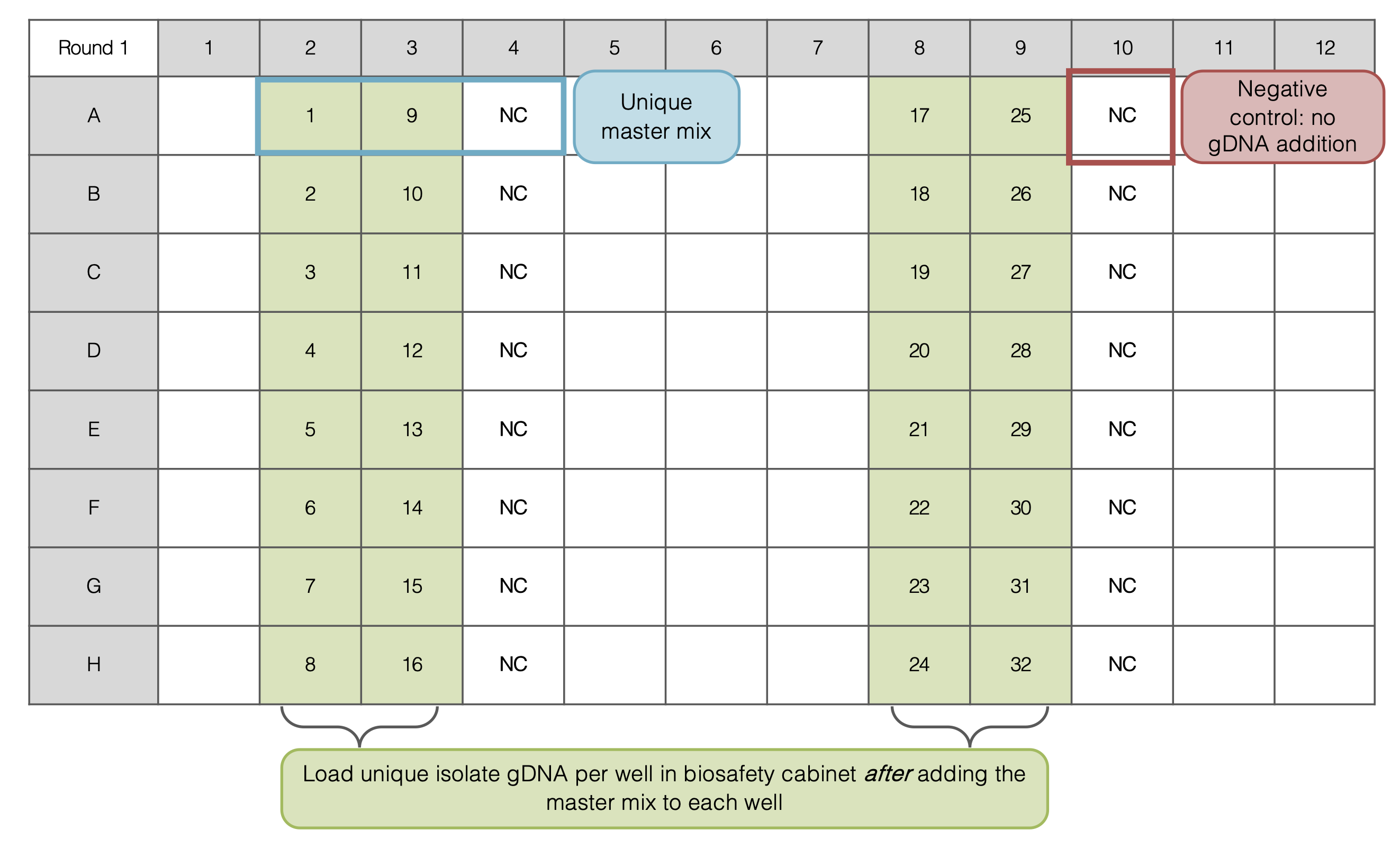
PCR Round 1: Plate Set Up
- For each PCR set up 32 samples (4 columns)
- Prepare 16 Master Mix (MM) solutions of “4x” per Multiplex Reaction (must have one negative control per MM)
- Add each reagent in the order listed above
- In each MM there will be two Primer sets (forward and reverse for each primer)
- Pipette 22 or 23 uL of each MM into the PCR plate as indicated below, then add gDNA in biosafety cabinet
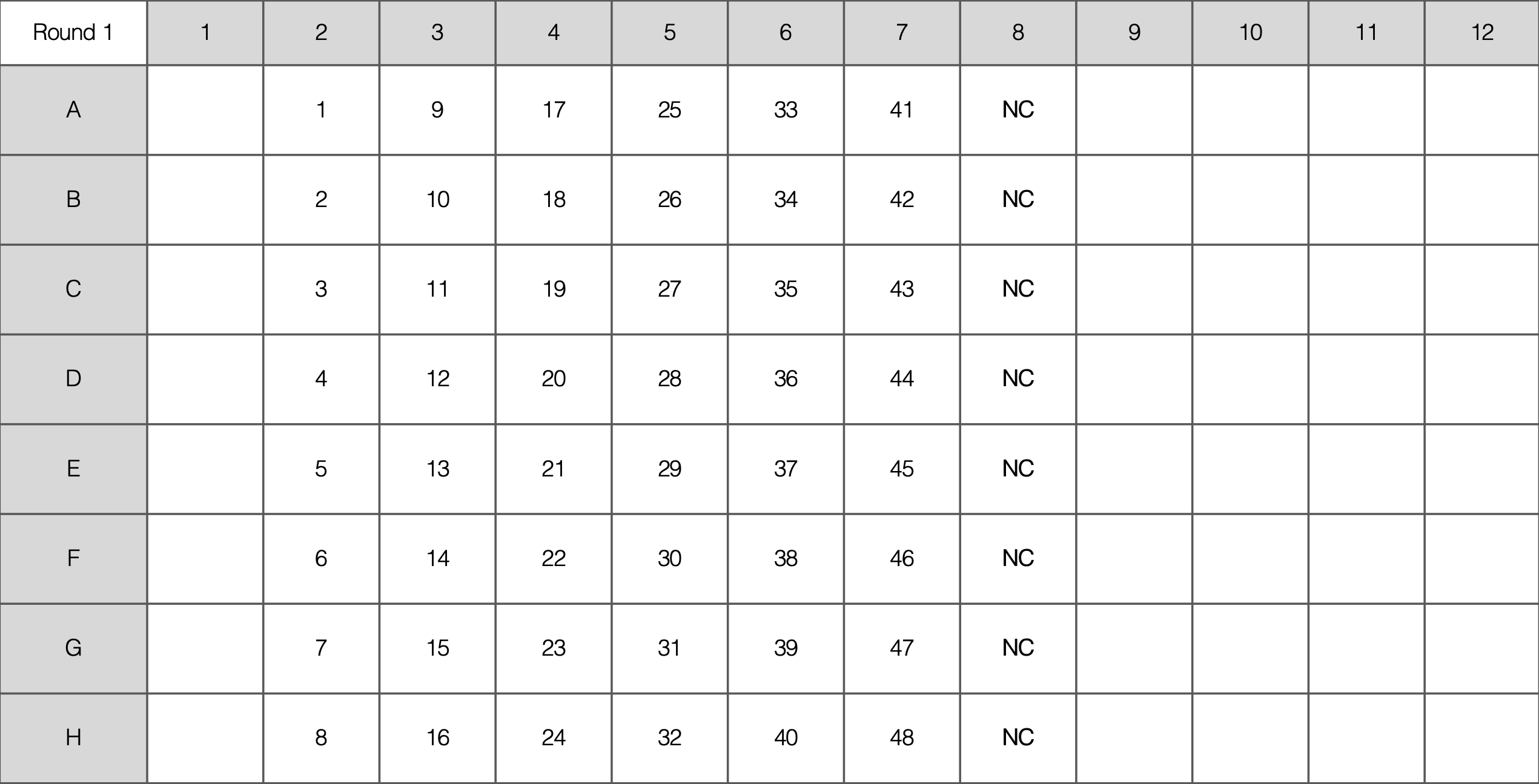
- For each PCR set up 48 samples (6 columns)
- Prepare 8 Master Mix (MM) solutions of “8x” per Multiplex Reaction (must have one negative control per MM)
- Add each reagent in the order listed above
- In each MM there will be two Primer sets (forward and reverse for each primer)
- Pipette 22 uL of each MM into the PCR plate as indicated below, then add gDNA in biosafety cabinet
PCR Round 2: Plate Set Up
- For each PCR set up 32 samples (4 columns)
- Prepare 16 Master Mix (MM) solutions of “4x” per Multiplex Reaction (must have one negative control and MID per MM)
- Add each reagent in the order listed above
- In each MM there will be two Primer sets (forward and reverse for each primer)
- Pipette 23 or 24 uL of each MM into the PCR plate as indicated below, then add gDNA in biosafety cabinet
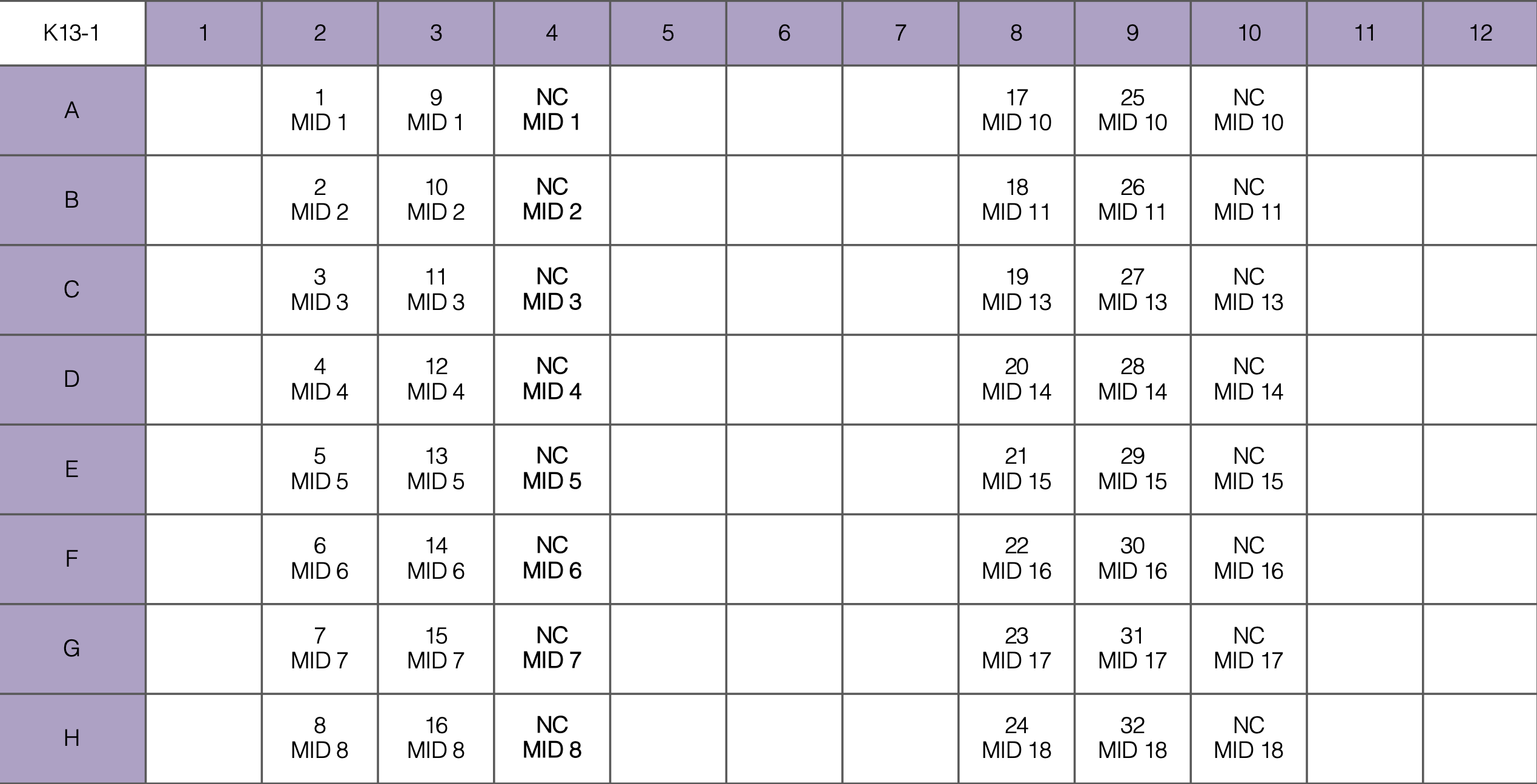
- For each PCR set up 48 samples (6 columns)
- Prepare 8 Master Mix (MM) solutions of “8x” per Multiplex Reaction (must have one negative control and MID per MM)
- Add each reagent in the order listed above
- In each MM there will be two Primer sets (forward and reverse for each primer)
- Pipette 23 or 24 uL of each MM into the PCR plate as indicated below, then add gDNA in biosafety cabinet
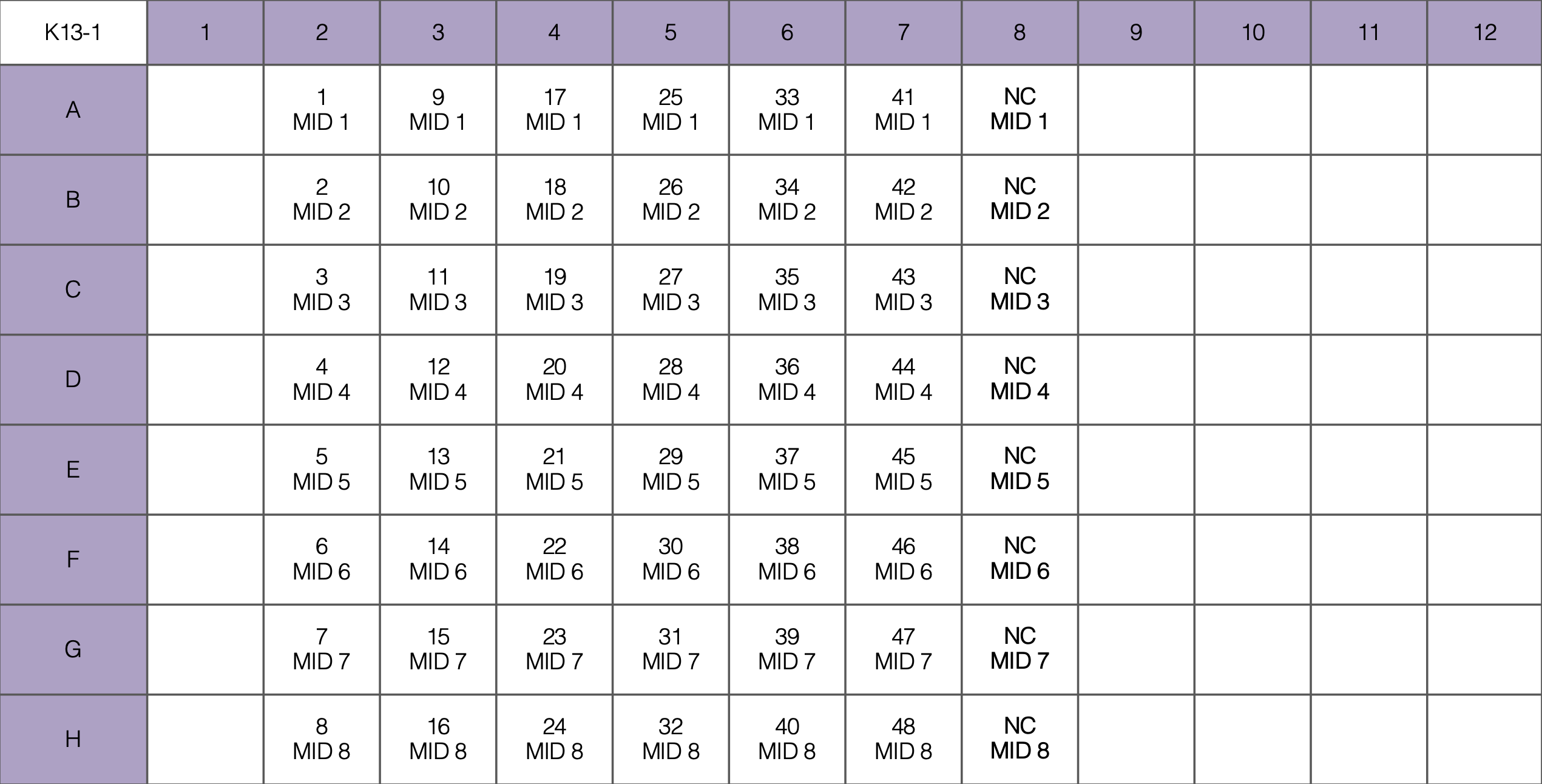
Gel Electrophoresis
Expected Bands
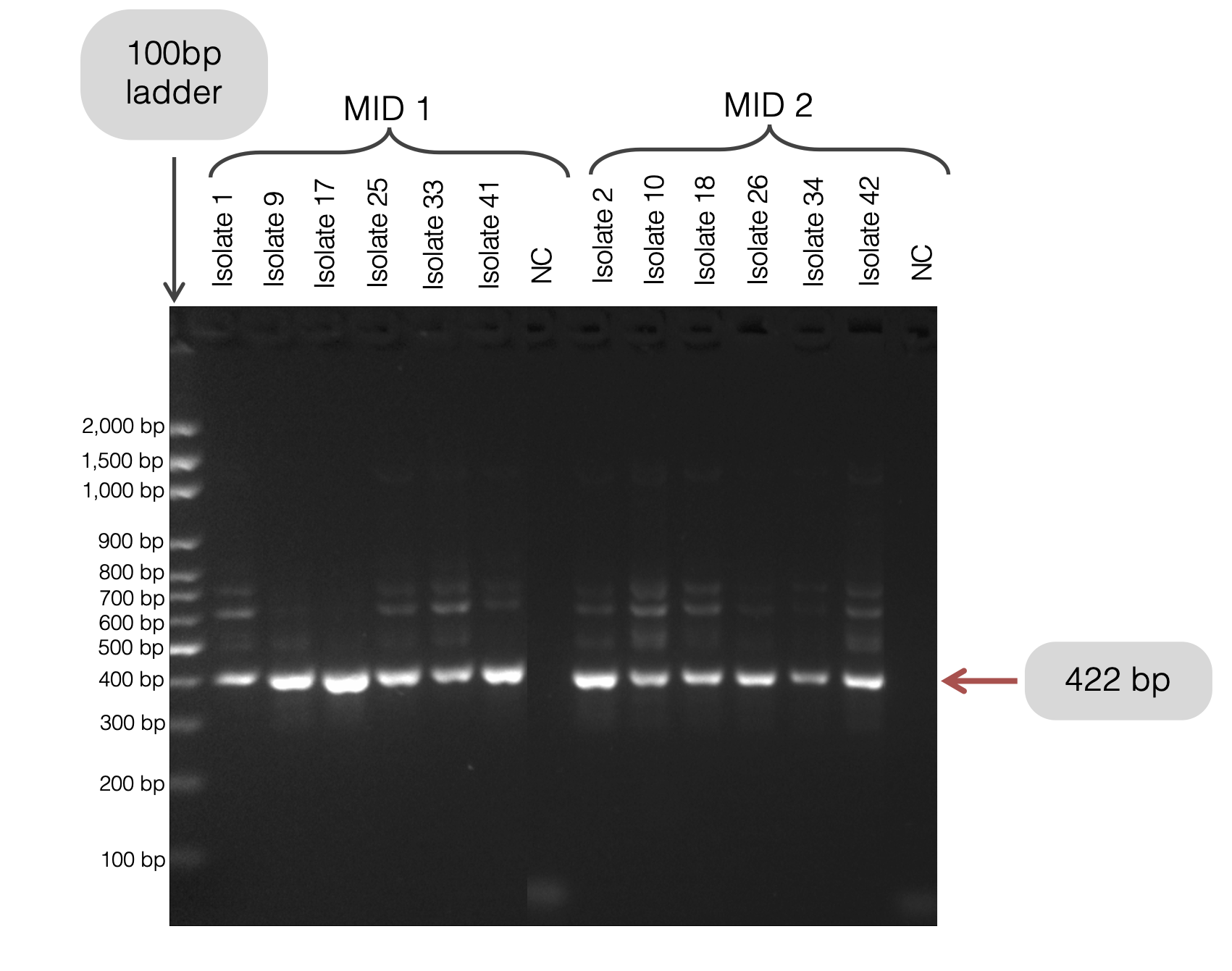
After every PCR Round 2, 5 μL per well of each sample was visualised by a gel electrophoresis. Run a 2% agarose gel at 100 V for 110 minutes EXCEPT for crt which should be run at 100 minutes (smaller fragment).
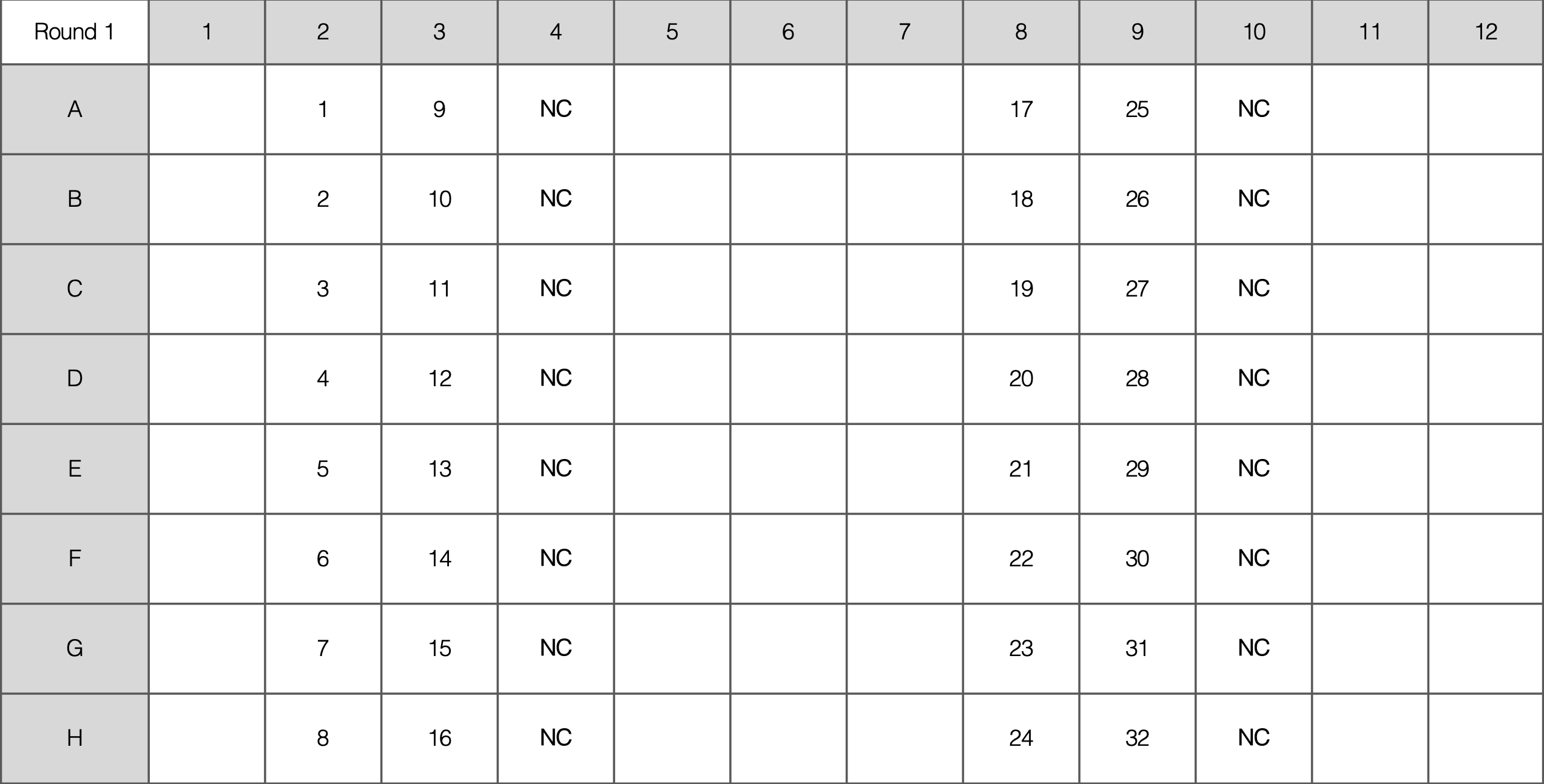
Expected Gel
For example, here are 18 isolates and 2 negative controls for K13-1:
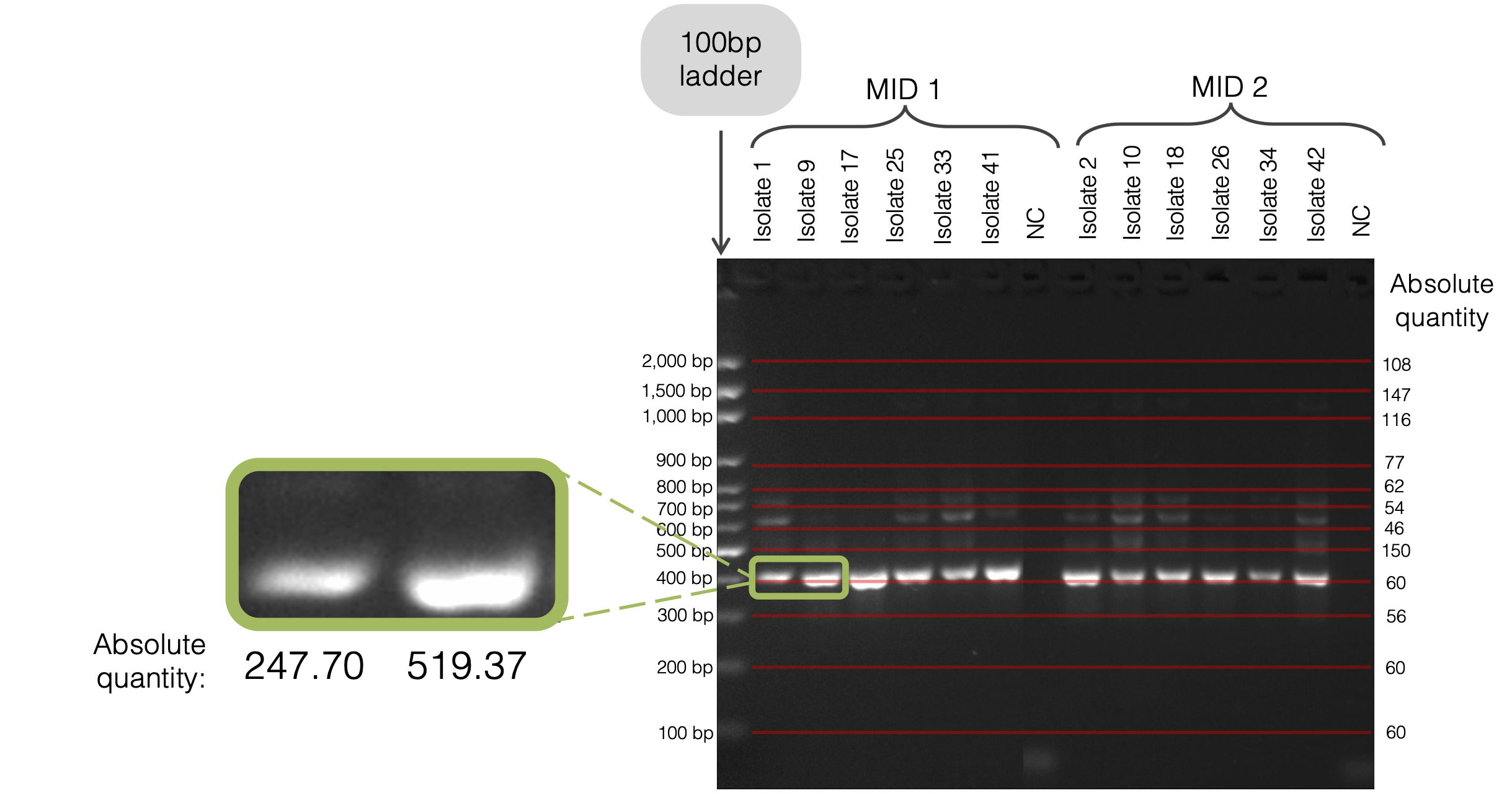
Agarose-gel densitometry
For each reaction, agarose-gel densitometry (EZQuant, Biorad, US) was used to quantify the DNA concentration of the fragment sizes using a 100bp ladder standard (Axygen, CA, US and MaestroGen, Taiwan).
Keep these tabulations and refer to Post-PCR: Prepare for Sequencing
Reagents and Equipment
Reagents:
- Nuclease-free water
- 5x Buffer
- MgCl2 (25 mM)
- dNTPs (10 mM)
- G2 Flexi Taq (5U/μL)
- 100bp ladder standard
Laboratory consumables:
- 96-well plates
- 1 mL centrifuge tubes
- 10 μL micropipette and tips
- 20 μL micropipette and tips
- 200 μL micropipette and tips
- Molecular Grade Agarose
Equipment:
- PCR Machine and agarose-gel densitometry capabilities: Gel Doc\(^{TM}\) EM System
- Electrophoresis Apparatus
Troubleshooting
Gel Electrophoresis
- Bands can be seen at the extreme bottom of the gel: primer dimers
- Bands can be seen at the round 1 product length:
- K13-out: 845 bp
- mdr1-out-1: 579 bp
- mdr1-out-2: 1,468 bp
- crt-out: 777 bp
- dhfr-out: 766 bp
- dhps-out: 1,189 bp
- aat1-out: 969 bp
- pfs47-out: 1,320 bp